Gene expression quantification for cancer biomarkers
암과 질병을 진단하기 위해서는 주로 조직학적 검사를 진행하는데, 이는 시간이 오래 소요될 뿐 아니라, 복잡하고 검사 비용이 높아 실제 질병으로 인한 증상이 확인된 환자들에게 적용되고 있다.
이상적인 진단 방법은 빠르고, 효율적이어야 하며, 증상이 나타나기 전에 낮은 비용으로 정확하게 환자를 선별할 수 있어야 한다. 이를 위해, 잠재적인 암과 질병의 biomarker로써 체액이나 FFPE 조직으로부터
추출한 핵산을 활용하고자 하는 연구가 이루어지고 있다. 임상적으로 이미 핵산은 산전 유전자 검사를 위해 이용되어 왔고, 다양한 질병 진단을 위해 수많은 biomarker들이 활용되고 있다. 이런 biomarker는
크게 DNA, mRNA, microRNA (miRNA), long noncoding RNA (lncRNA) 4 분류로 나누어 볼 수 있다.
간암, 유방암, 전립선암과 같은 다양한 종류의 암에서 mRNA marker가 보고되고 있다. 하지만, mRNA는 가변성과 변형이 높아 정확한 진단에 적용하기에는 어려움이 있다. miRNA는 보통 ~20 nt 정도의 크기를
가지는 noncoding RNA로, 유전자 발현을 조절하는 것으로 알려져 있으며, coding mRNA와는 다르게 비교적 안정적 형태를 지닌다. 현재까지 수천개의 unique miRNA가 확인되었으며, 수많은 종류의 암과
질병에 영향을 미치는 것으로 확인되었다. IncRNA는 보통 200 nt 이상의 긴 전사체로, 하나의 특정 단백질을 코딩하지 않고 유전자 발현을 조절하는 것으로 보이며, 다양한 종류의 암에서 수십만개의 IncRNA가 확인되었다.
PCR, qPCR, RT-qPCR은 이러한 다양한 종류의 샘플로부터 유전자 발현을 확인하고 정량하기 위해 사용되는 분자적 기법으로, 빠르고 정확하며, 민감도 있게 cancer biomarker를 확인할 수 있다.
Highlighted products - Whole-genome SNP array with the Titanium DNA Amplification Kit
Cytogenetic microarray는 저비용으로 빠르게 특정 유전자의 변형을 확인할 수 있는 방법으로, 저발현 mosaicism과 heterozygosity의 손실, copy number 변화 등을 확인할 수 있다 (Eldai
et al.
2013). 이러한 유전자 변형은 다양한 종류의 암의 진단과 치료에 영향을 미치는 것으로 알려져 있다.
다카라바이오는 다양한 암의 유전자 변형을 확인하기 위한 microarray에 사용할 수 있는
Titanium® DNA Amplification Kit를 지원하고 있다. 이를 이용해, Weyant
et al. 는 종양에서
BRAF와 같은 유전자 변형을 확인하였으며, Lopes
et al. 는
초기 단계의 급성림프구백혈병에서
IKFZ1 결실을 확인하였다.
Titanium® Polymerase를 Affymetrix GeneChip Mapping 500K Assay의 SNP genotyping에
사용하면, 높은 yield와 재현성으로 최고 수준의 결과를 얻을 수 있다 (그림 1).
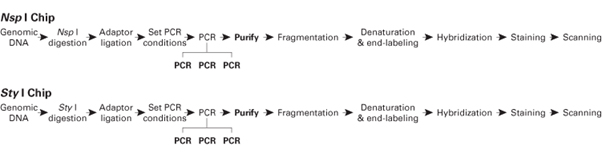 그림 1. Overview of the GeneChip Mapping 500K Assay.
Highlighted products - Quantitative detection of gene expression using RT-qPCR and qPCR kits
그림 1. Overview of the GeneChip Mapping 500K Assay.
Highlighted products - Quantitative detection of gene expression using RT-qPCR and qPCR kits
RT-qPCR과 qPCR은 circulating tumor cell과 같은 다양한 샘플로부터 획득한 RNA, gDNA의 유전자 발현을 민감도 있게 정량 분석 할 수 있다 (Lianidou
et al. 2016).
다카라바이오는 연구자가 원하는 제품을 선택할 수 있도록, 다양한 형태의 qPCR 제품을 지원하고 있다. 이를 이용하면 암연구를 위해 빠르고 정확하며 민감도 높게 유전자를 검출 할 수 있다.
FFPE 조직과 같이 귀중한 샘플에 적용할 수 있도록,
Prelude™ PreAmp Master Mix를
판매하고 있으며, 이를 사용하면 12.5 pg의 극소량의 DNA로부터 편향성 없는 분석이 가능하다.
본 분석 방법을 이용해, Makino
et al. 는 자궁 평활근 육종 (uterine leiomyosarcoma)의 FFPE 샘플과 동결보관된 샘플에서 TrkB signaling의 변화를 확인할 수 있었다. Chen
et al. 는
유방암의 FFPE 샘플 55개로부터 key miRNA를 식별하였으며, Li
et al. 는 인간 간세포암 종양 조직에서
MAF1을 확인하였다. Jiang
et al. 는 전립선암 조직과 세포로부터
SARI 종양 억제 유전자 발현 감소로 인해 이를 대체하는
RSK4 발현이 증가함을 정량하여 확인하였으며, FFPE 조직 샘플에서 유방암과 관련한 유전자 162개가 재현성 높게 발현함을 확인하였다.
-
FFPE 조직에서 편향성 없는 Preamplification (클릭)
Highlighted products - Sensitive miRNA quantification with Mir-X™ miRNA qRT-PCR TB Green® kits
특이한 miRNA를 정확하고 민감도 있게 정량분석 하기 위해서는 다양한 암의 signaling network를 이해하고 확인하는 것이 중요하다 (Hayes
et al. 2014).
다카라바이오는
Mir-X™ miRNA qRT-PCR TB Green® kits를
이용해 single-tube 내 single step으로 miRNA의 1st strand cDNA를 합성하고, miRNA-specific primer와
Advantage qPCR chemistry를 이용하여, miRNA를 특이적으로 정량 분석할 수 있도록 지원하고 있다 (그림 2). 이를 이용하면, 혈장 (Kanemaru
et al. 2011), exosome (Lee
et al. 2013) 및
FFPE 조직 (Lee
et al. 2013)을 포함한 수많은 종류의 샘플에서 50 copies의 miRNA까지 간단하고 정확하게 검출할 수 있다.
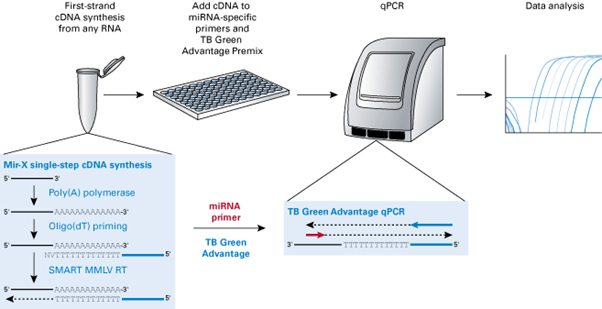 그림 2. Mir-X™ miRNA qRT-PCR TB Green® kit workflow
Highlighted products - High-throughput gene expression analysis with the SmartChip™ Real-Time PCR System
그림 2. Mir-X™ miRNA qRT-PCR TB Green® kit workflow
Highlighted products - High-throughput gene expression analysis with the SmartChip™ Real-Time PCR System
Biomarker 연구의 어려움 중 하나는 다양한 종류의 샘플에서 수백개의 후보 마커를 스크리닝 해야 한다는 점이다. 많은 타겟을 빠르고 민감하게 분석하기 위해, 다카라바이오는
대량 (High-throughput) qPCR을 위한
SmartChip™ System을
지원하고 있다. 이는 혈액, 세포주, T cell, 혈장 등 다양한 종류의 샘플로부터 암, 질병의 miRNA (Choo
et al. 2014), lncRNA (Leucci
et al. 2016), mRNA (Chen
et al. 2016)를
확인하는 데 활용되었다. 이러한 연구에서
SmartChip™ System은
수많은 분석과 샘플 확인을 위해 사용되었으며, 각각의 질병이 가지는 고유한 biomarker와 그 변화를 확인할 수 있었다.
다카라바이오의
SmartChip™ System를 질병 고유의 biomarker를
대량 분석에 이용함으로써 mRNA, miRNA, IncRNA 분석용 panel을 디자인하고 활용할 수 있다.
-
다양한 biomarker panel과 연구 (클릭)
 그림 3. SmartChip™ Real-Time PCR System
[원문] Gene expression quantification for cancer biomarkers
[참고문헌]
그림 3. SmartChip™ Real-Time PCR System
[원문] Gene expression quantification for cancer biomarkers
[참고문헌]
- Chen, X. et al. The role of miRNAs in drug resistance and prognosis of breast cancer formalin-fixed paraffin-embedded tissues. Gene 595, 221-226 (2016).
- Chen, X. et al. Defining a population of stem-like human prostate cancer cells that can generate and propagate castration-resistant prostate cancer. Clin. Cancer Res. 22, 4505-4516 (2016).
- Choo, K. B. et al. MicroRNA-5p and -3p co-expression and cross-targeting in colon cancer cells. J. Biomed. Sci. 21:95 (2014).
- Eldai, H. et al. Novel genes associated with colorectal cancer are revealed by high resolution cytogenetic analysis in a patient specific manner. PLoS One 8, e76251 (2013).
- Hayes, J., Peruzzi, P. P. & Lawler, S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 20, 460-469 (2014).
- Jiang, Y. et al. Aberrant expression of RSK4 in breast cancer and its role in the regulation of tumorigenicity. Int. J. Mol. Med. 40, 883-890 (2017).
- Kanemaru, H. et al. The circulating microRNA-221 level in patients with malignant melanoma as a new tumor marker. J. Dermatol. Sci. 61, 187-193 (2011).
- Lee, J.-K. et al. Exosomes derived from mesenchymal stem cells suppress angiogenesis by down-regulating VEGF expression in breast cancer cells. PLoS One 8, e84256 (2013).
- Lee, T. S. et al. Aberrant microRNA expression in endometrial carcinoma using formalin-fixed paraffin-embedded (FFPE) tissues. PLoS One 8, e81421 (2013).
- Leucci, E. et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 531, 518-522 (2016).
- Li, Y. et al. MAF1 suppresses AKT-mTOR signaling and liver cancer through activation of PTEN transcription. Hepatology 63, 1928-42 (2016).
- Lianidou, E. S. Gene expression profiling and DNA methylation analyses of CTCs. Mol. Oncol. 10, 431-442 (2016).
- Lopes, B. A. et al. COBL is a novel hotspot for IKZF1 deletions in childhood acute lymphoblastic leukemia. Oncotarget 7, 53064-53073 (2016).
- Makino, K. et al. Inhibition of uterine sarcoma cell growth through suppression of endogenous tyrosine kinase B signaling. PLoS One 7, e41049 (2012).
- Weyant, G. W. et al. BRAF mutation testing in solid tumors: a methodological comparison. J. Mol. Diagn. 16, 481-485 (2014).







